Projects
Cell2Sentence: scaling Large Language Models for Next-Generation Single-Cell Analysis
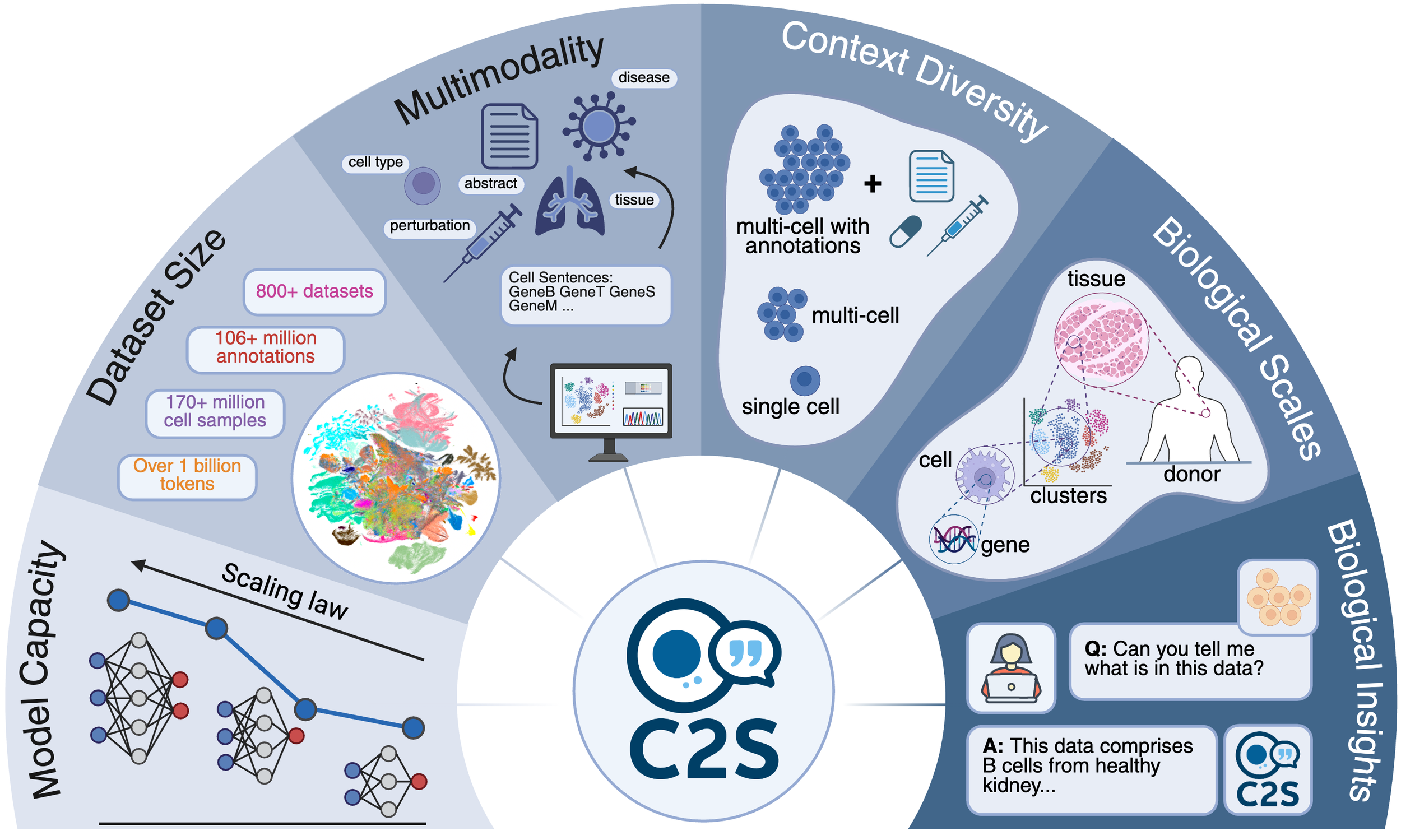
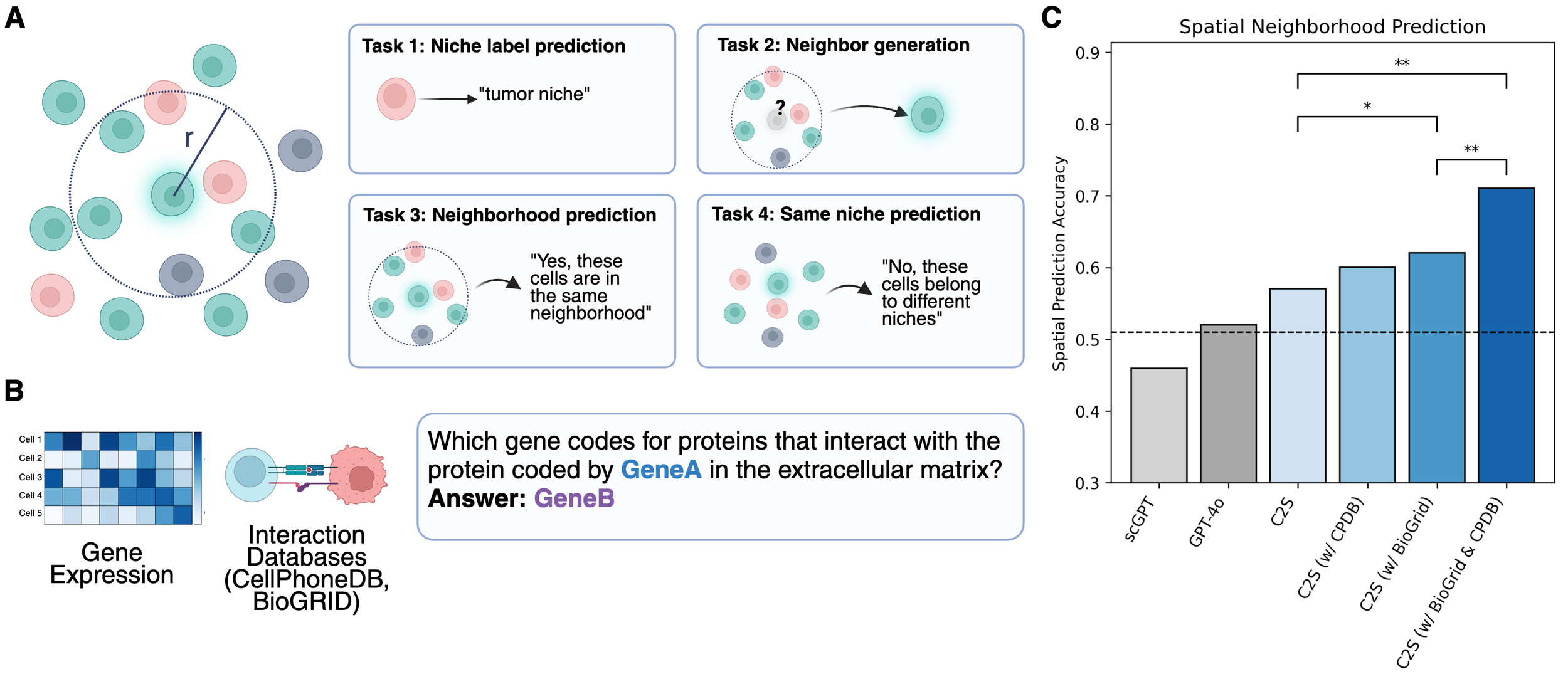
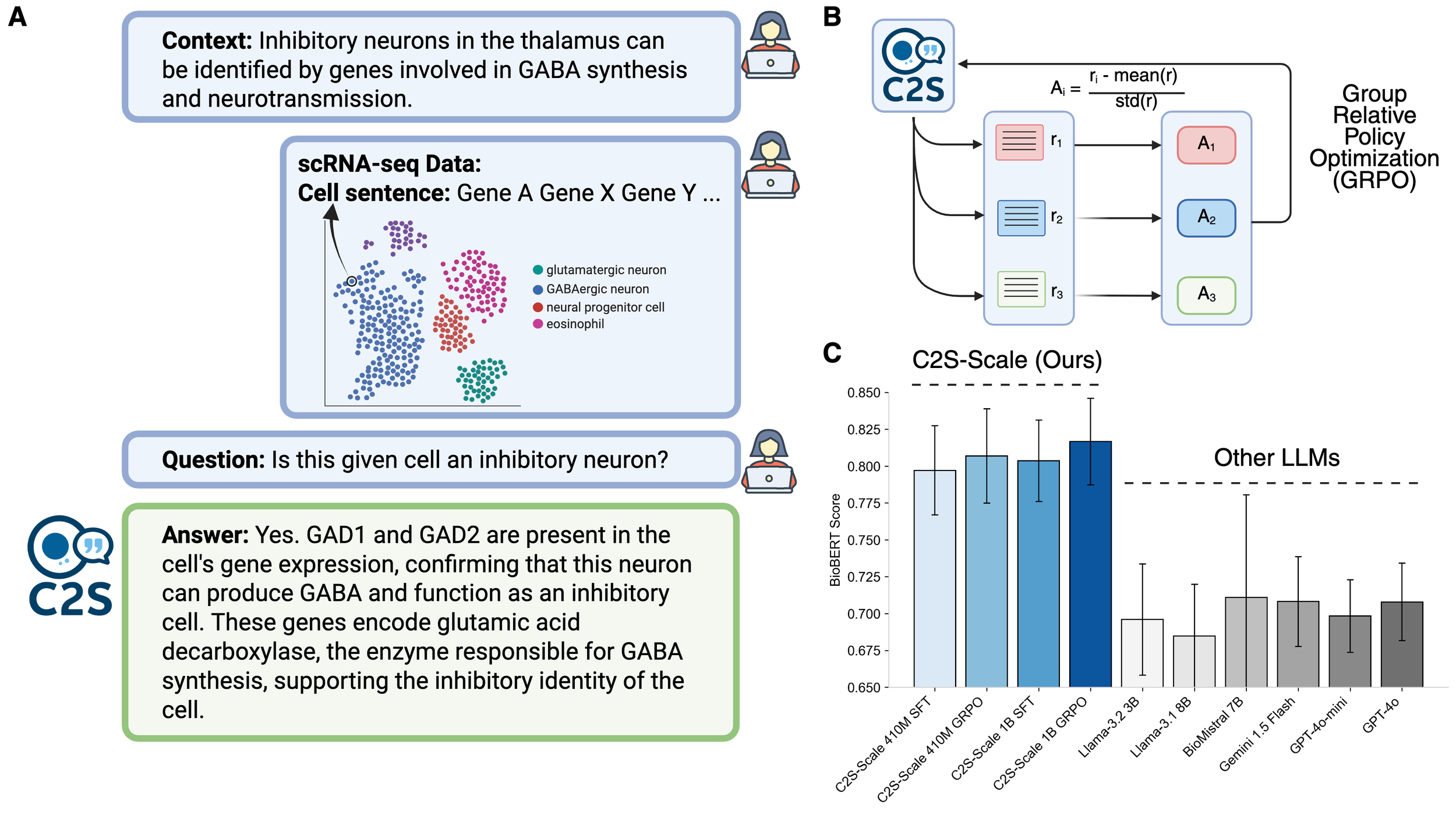
C2S is a family of large language models trained on over one billion tokens of single‑cell transcriptomic data, biological text, and metadata, to unify gene expression and natural language for single‑cell analysis. It delivers state‑of‑the‑art performance across predictive and generative tasks (e.g., perturbation response prediction, question answering, spatial reasoning) and introduces reinforcement‑learning‑based fine‑tuning for single-cell foundation models. By bridging transcriptomic and textual modalities at scale, C2S‑Scale paves the way for “virtual cells” and next‑generation foundation models in biology.






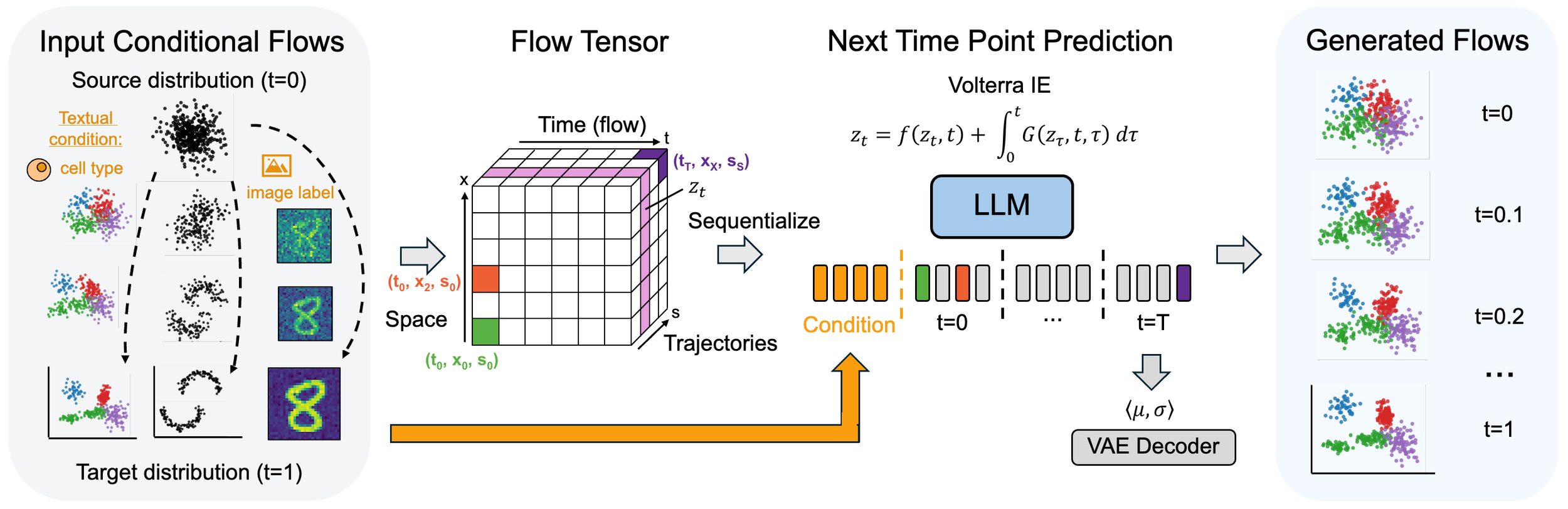
CaLMFlow: Volterra Flow Matching Using Causal Language Models
We introduce CaLMFlow, a novel framework that recasts ODE-based flow matching as a Volterra integral equation (VIE) while leveraging causal language models (CLMs) for continuous data generation. By discretizing both time and space, CaLMFlow transforms continuous trajectories into a sequence-based representation, allowing standard LLM architectures to capture long-range dependencies and flexibly incorporate textual prompts.
Caddi: Non-Markovian Discrete Diffusion with Causal Language Models
CaDDi introduces a non‑Markovian discrete diffusion framework that fuses the temporal trajectory of diffusion with left‑to‑right causal language modeling, so a single transformer can condition on multiple noisy timesteps while still leveraging pretrained LLM weights. This unified approach enables flexible, bidirectional sequence generation (e.g., text infilling) and, across natural‑language and protein tasks, it outperforms prior discrete diffusion baselines—narrowing the quality gap to large autoregressive transformers.
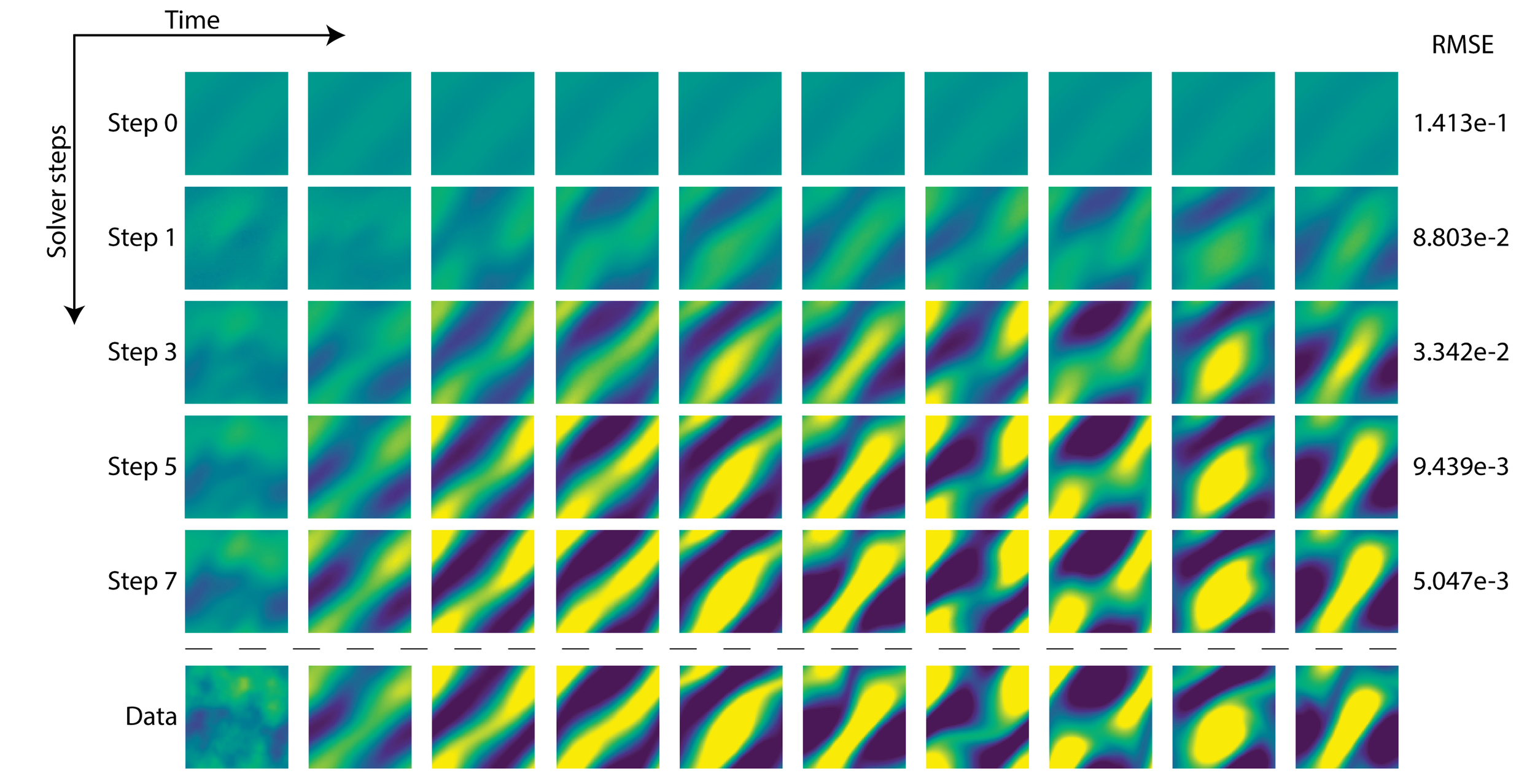
COAST: Intelligent Time-Adaptive Neural Operators
COAST introduces a novel neural operator learning framework that leverages a causal language model to dynamically adapt time steps when modeling dynamical systems governed by partial differential equations (PDEs). Our method intelligently predicts both the system's future states and optimal step sizes, effectively balancing computational efficiency and accuracy. We show that COAST consistently outperforms existing state-of-the-art approaches across various complex benchmarks, highlighting its ability to efficiently handle continuous-time problems by adapting to intrinsic system dynamics.
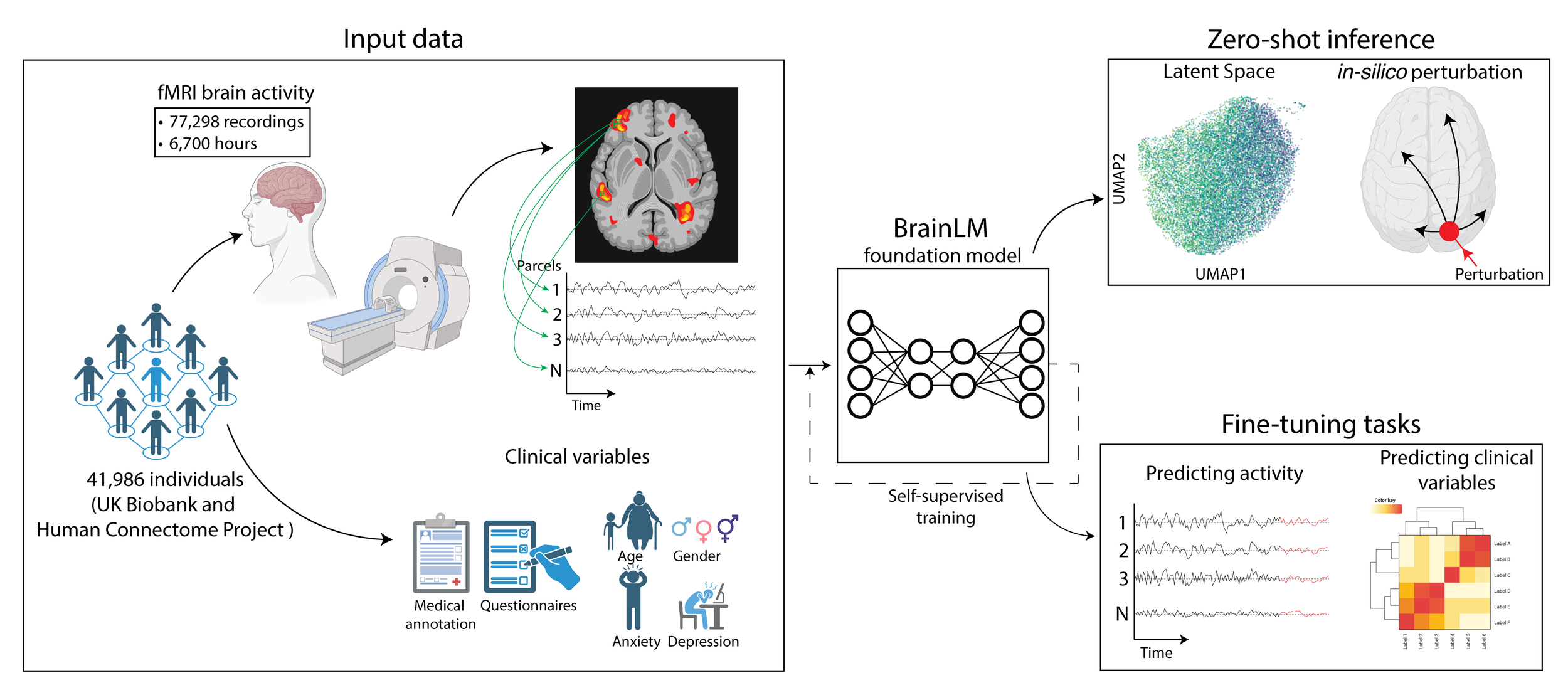
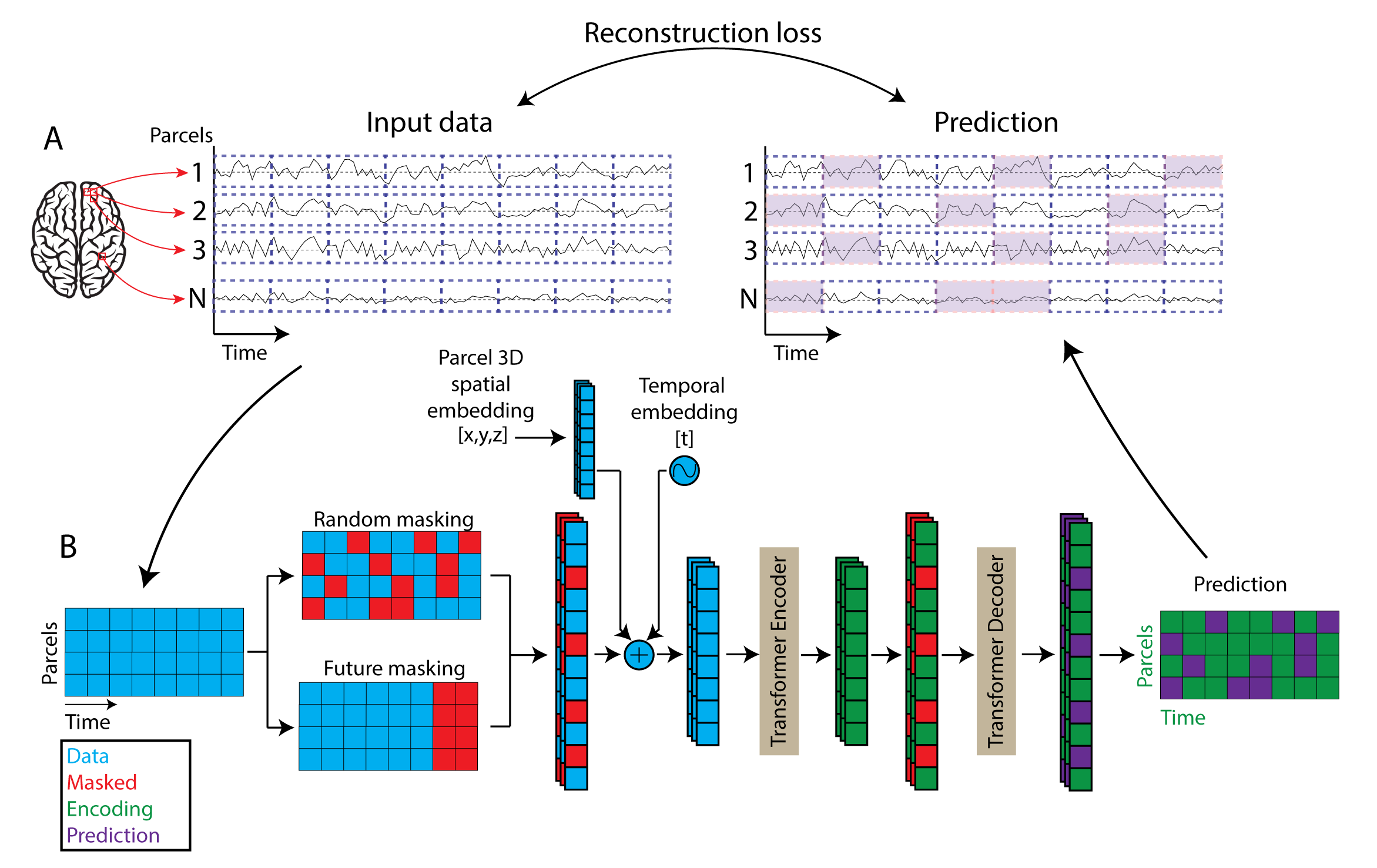
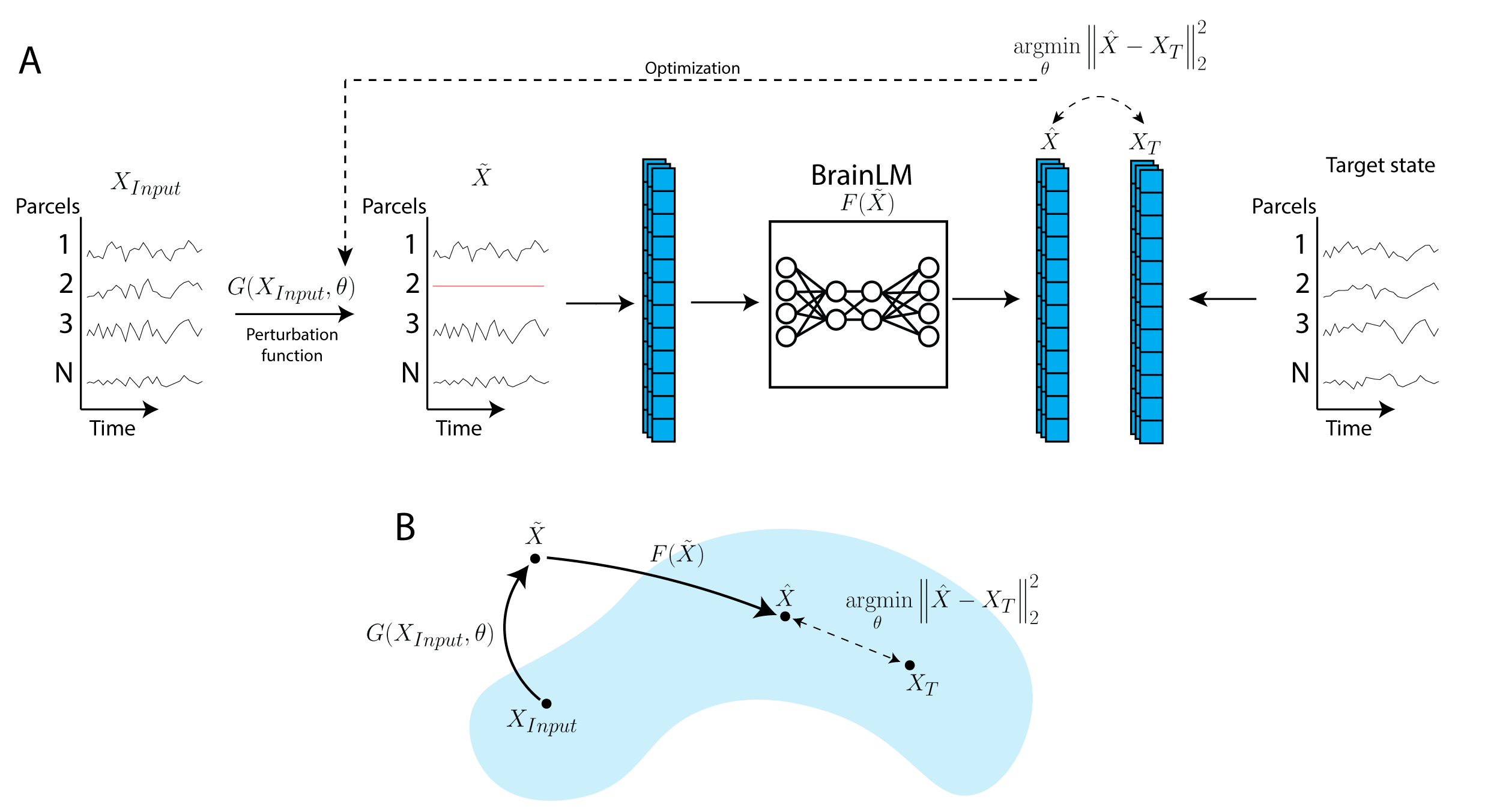
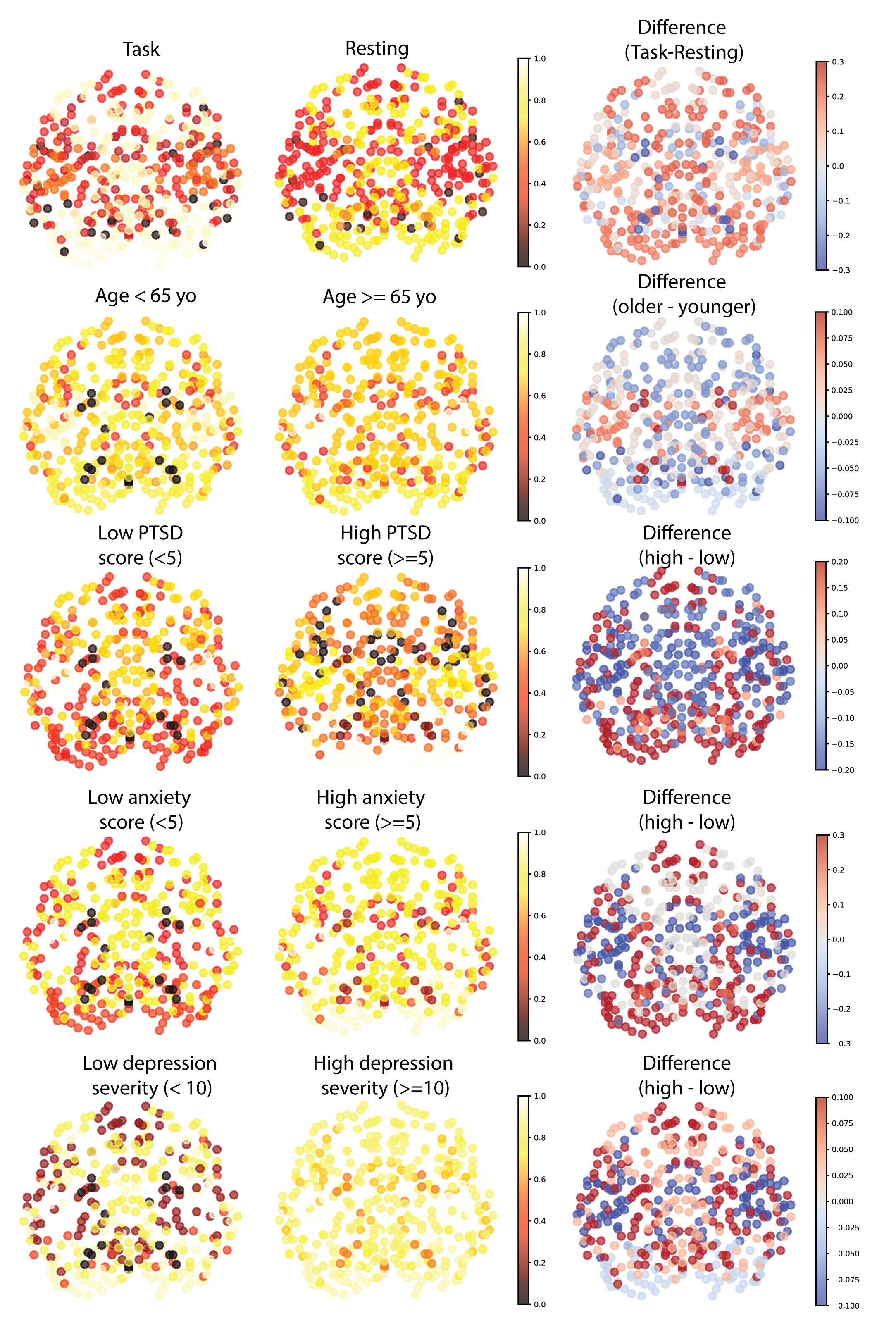
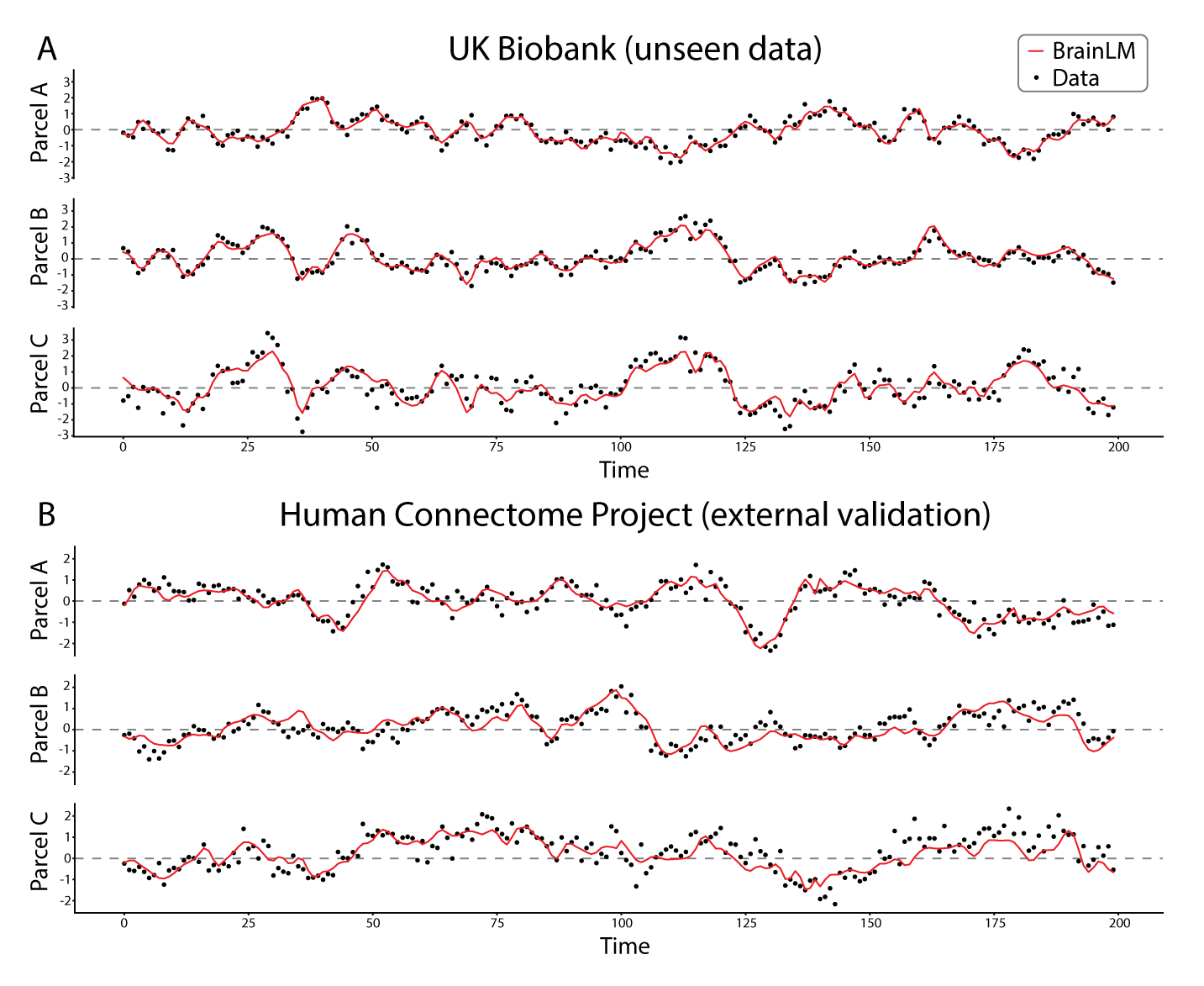
BrainLM: A Foundation Model for Brain Activity Recordings
BrainLM is a self‑supervised foundation model trained on 6,700 hours of fMRI data, excelling at clinical prediction tasks and zero‑shot network identification. A novel prompting technique lets it simulate how brain activity changes under different conditions.
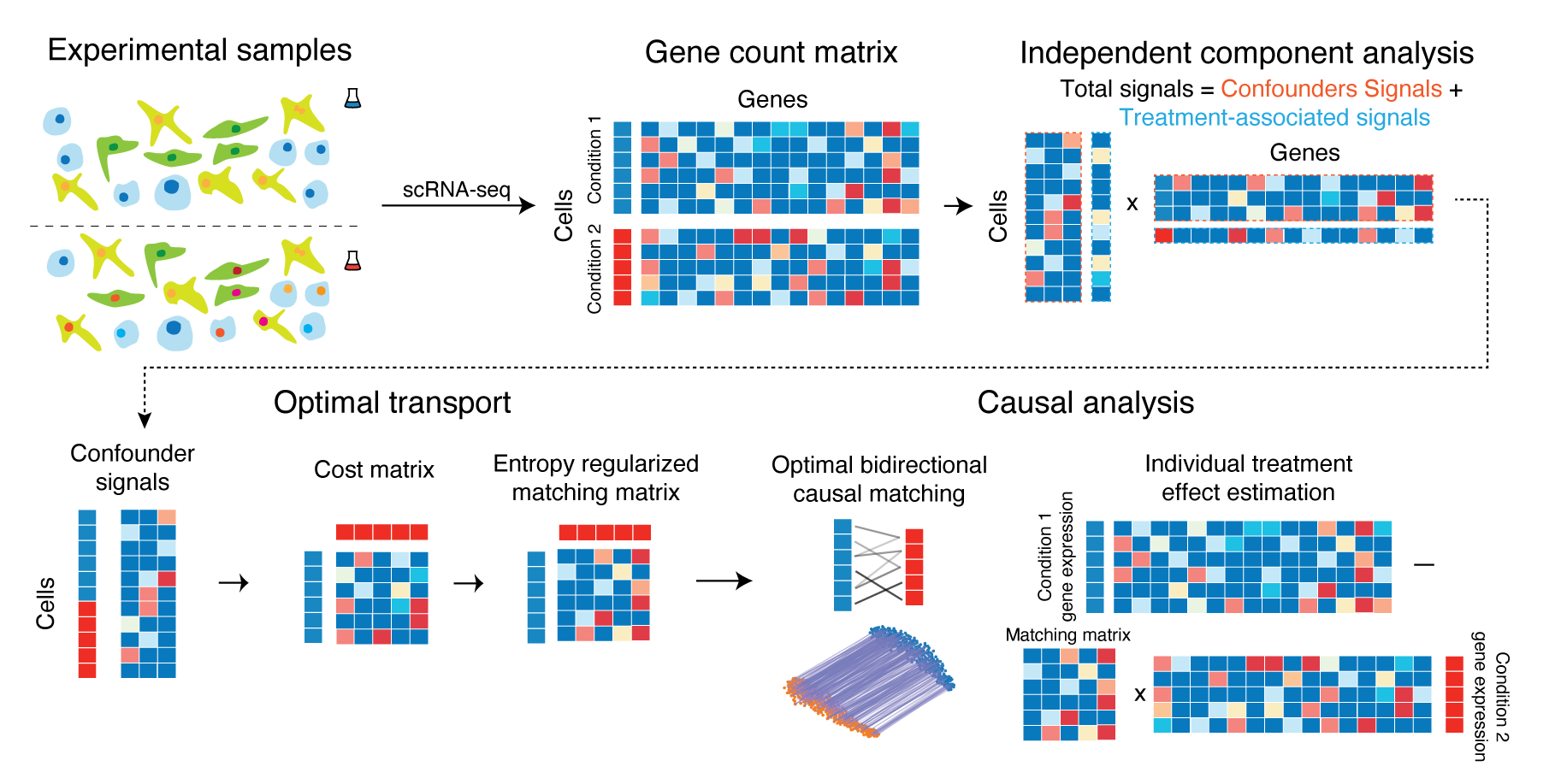
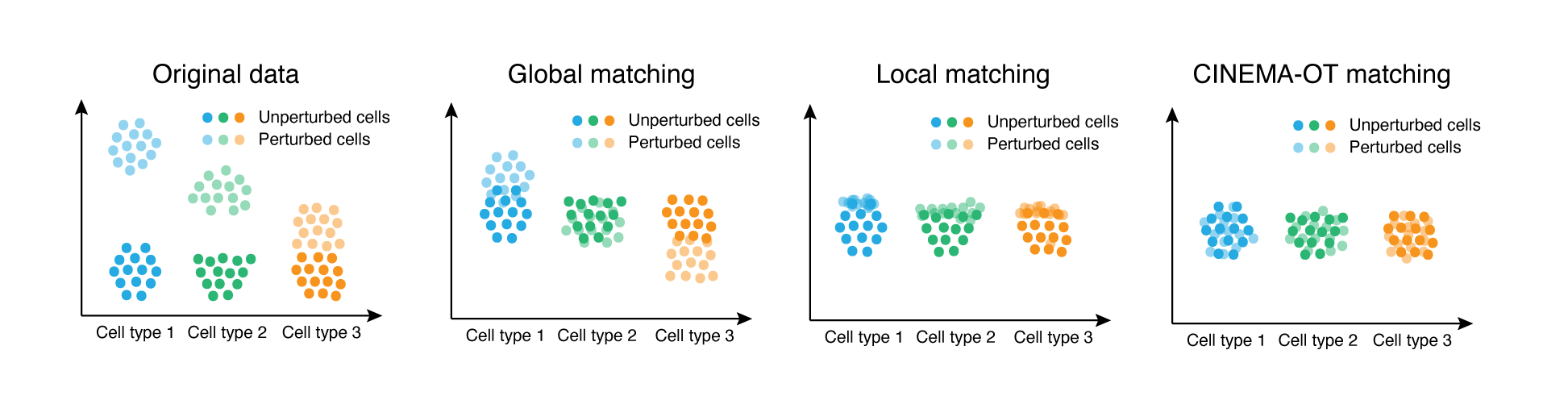
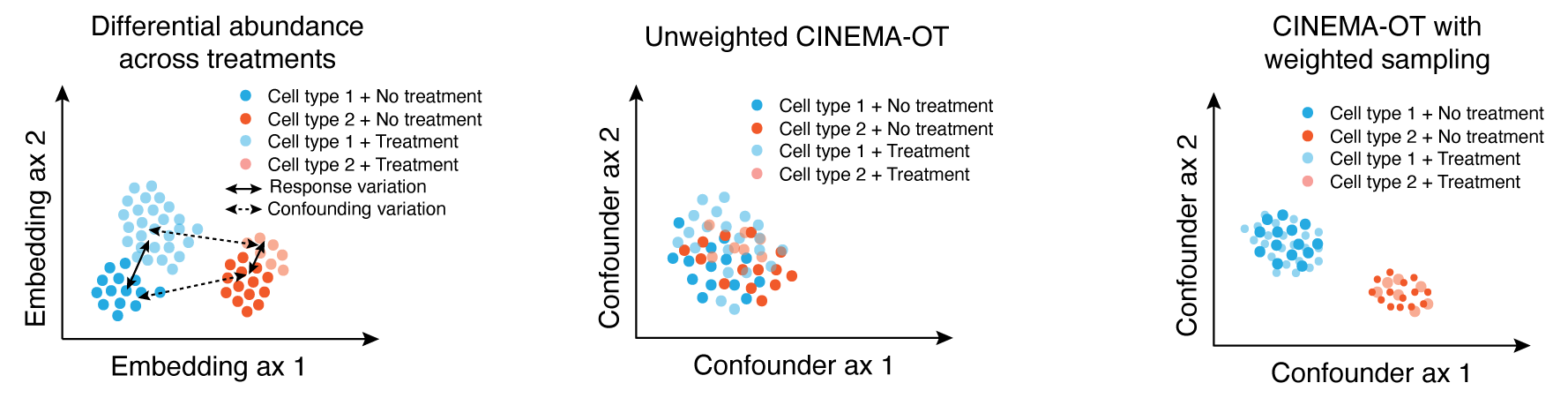
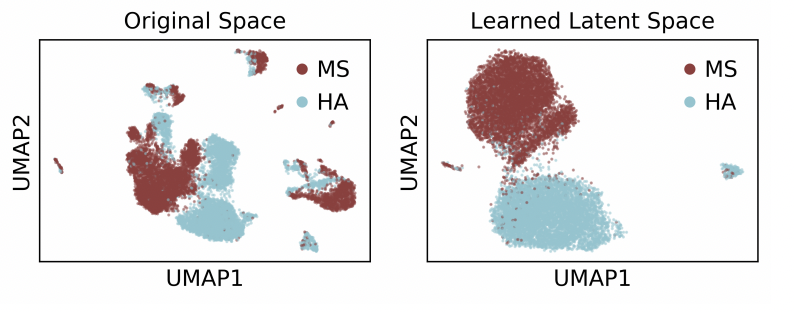
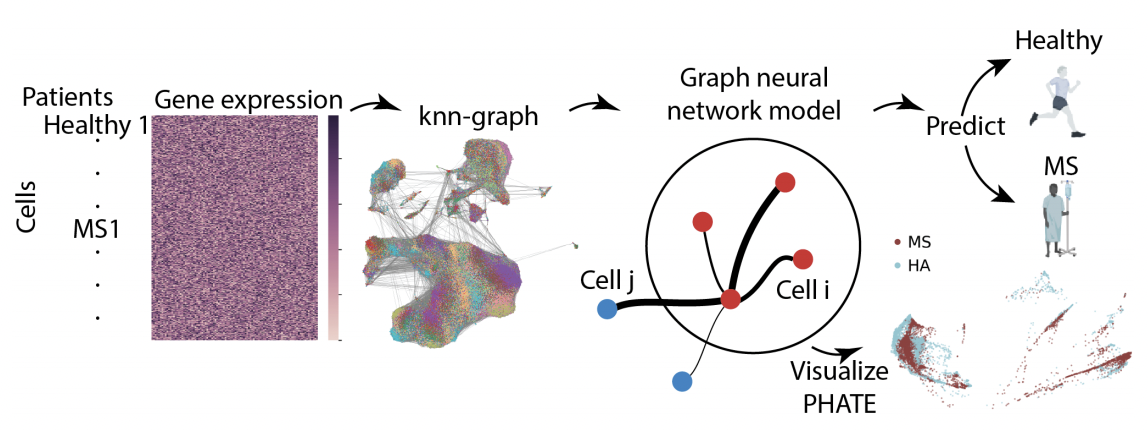
Causal identification of single-cell experimental perturbation effects with CINEMA-OT
CINEMA‑OT is a causal‑inference tool that creates counterfactual cell pairings to disentangle true treatment effects from confounders in single‑cell perturbation data. It outperforms existing methods on both simulated and real datasets and has revealed new antiviral‑response mechanisms in airway organoids and immune cells.
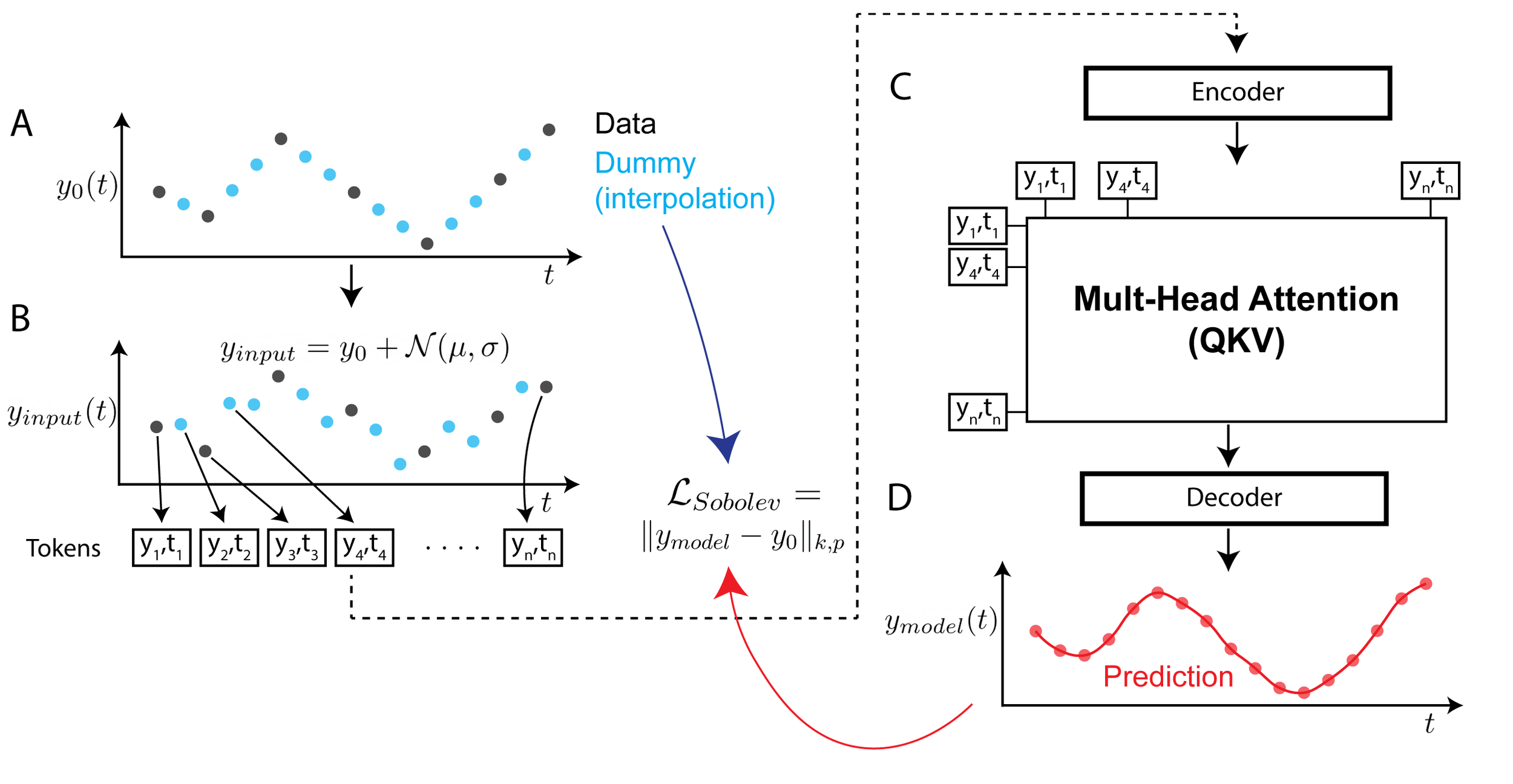
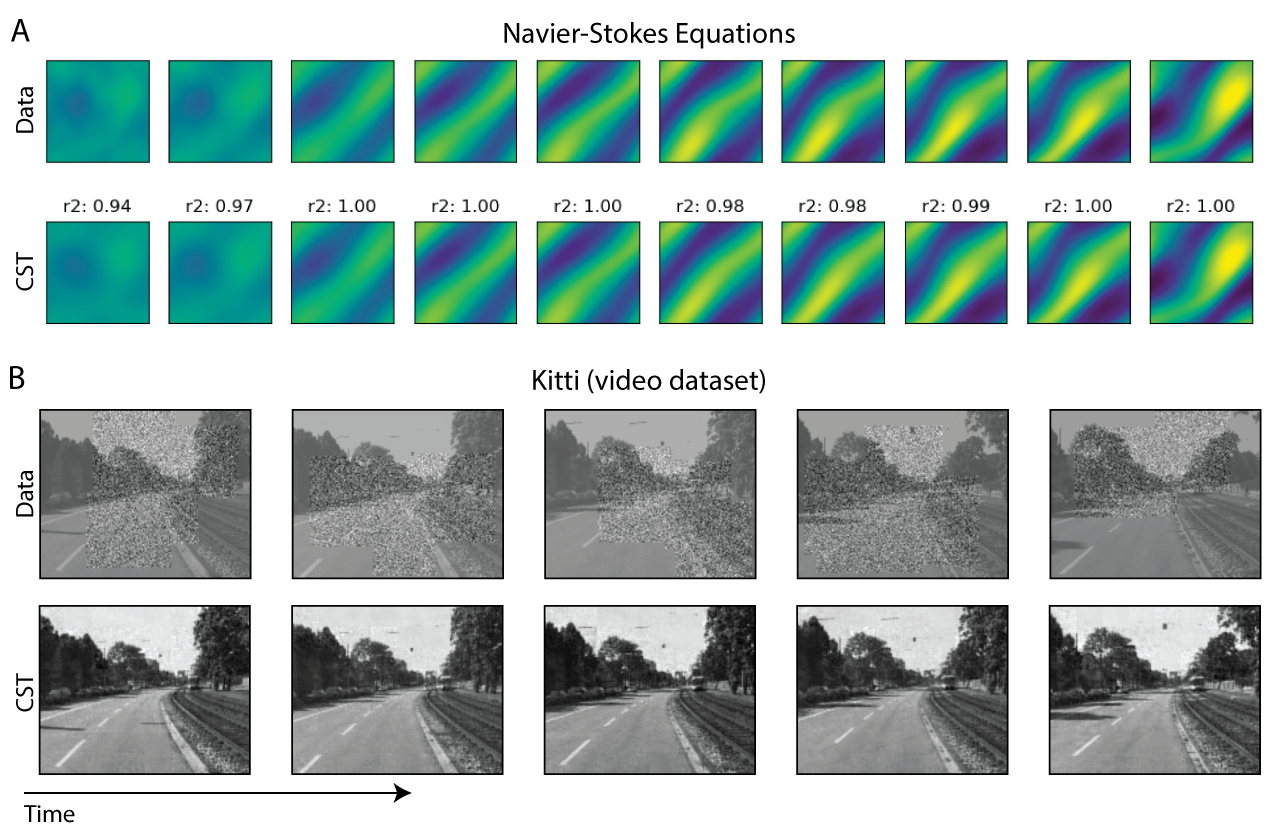
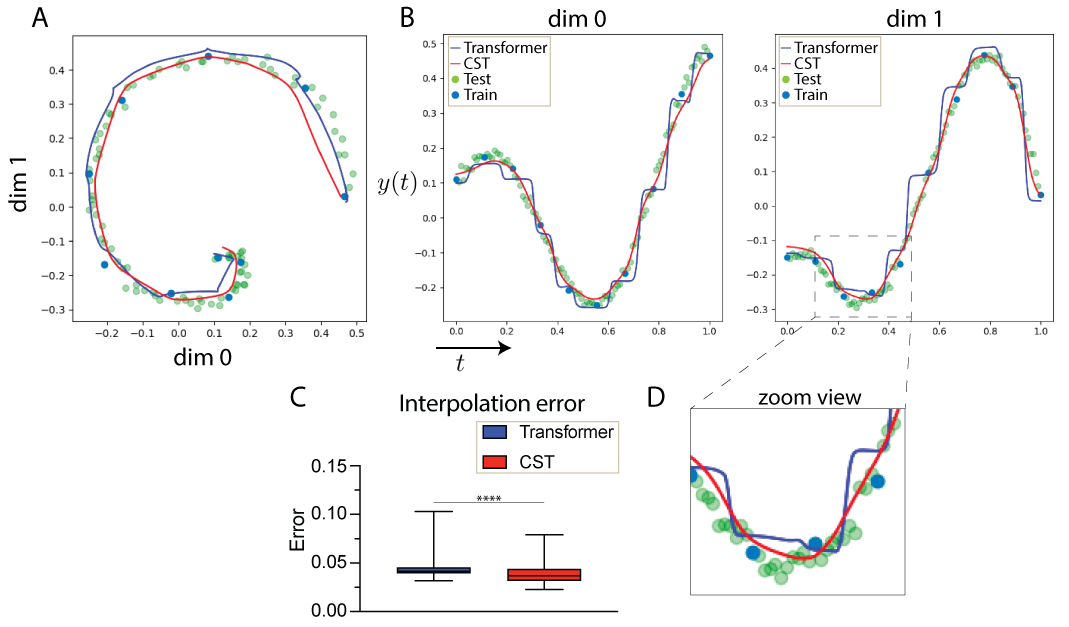
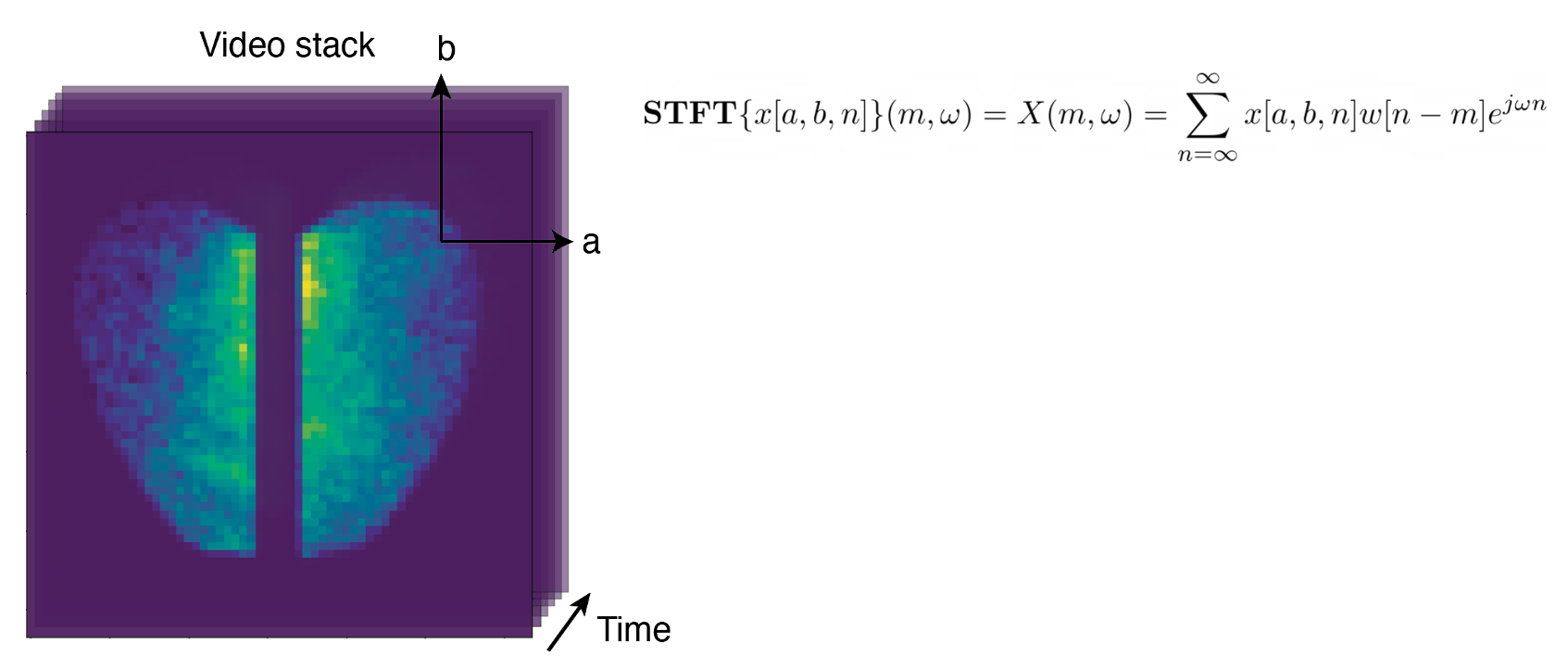
Continuous Spatiotemporal Transformers
Standard transformers have limitations in modeling continuous data due to their discrete nature. CST extends transformers to continuous data by adding a Sobolev‑based regularization that enforces smooth outputs, enabling accurate interpolation of irregular, noisy time‑series. It outperforms transformers, neural ODEs, and other baselines on both synthetic and real spatiotemporal datasets—including whole‑cortex calcium imaging.
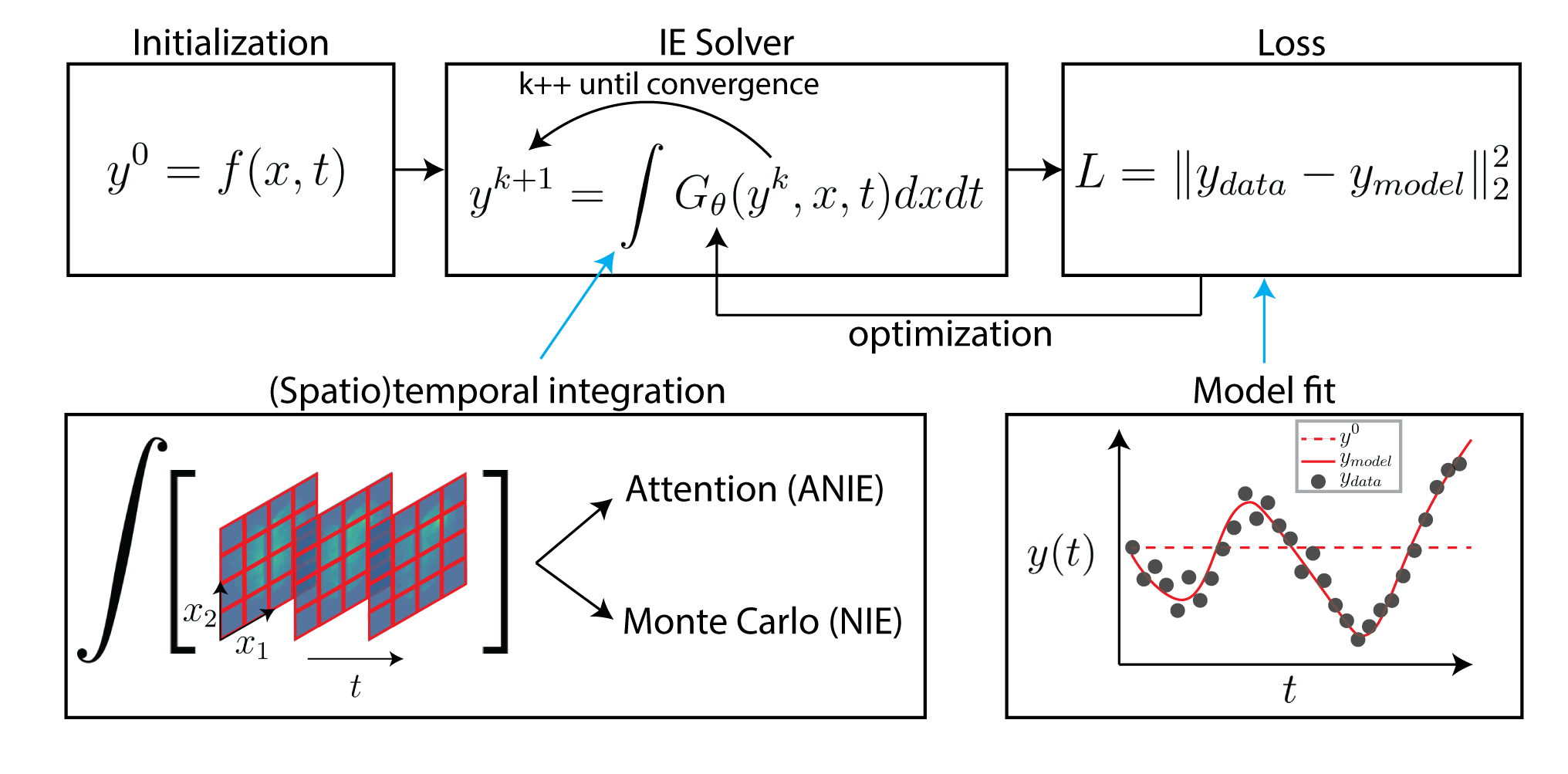
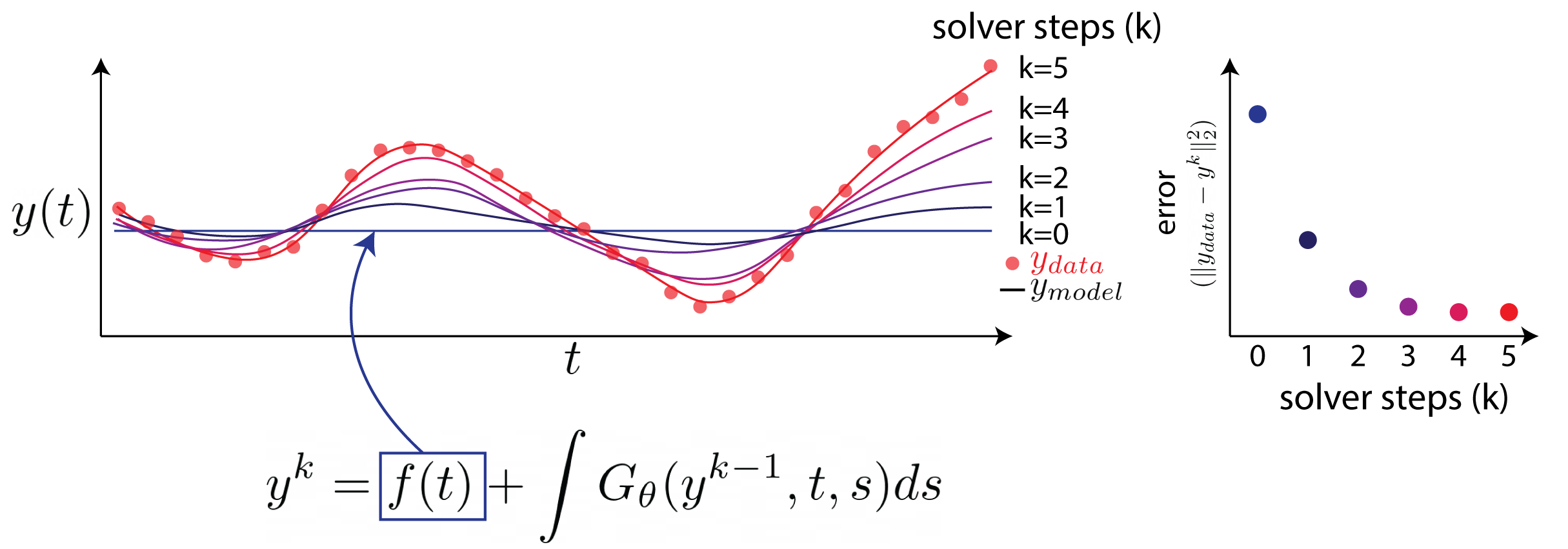
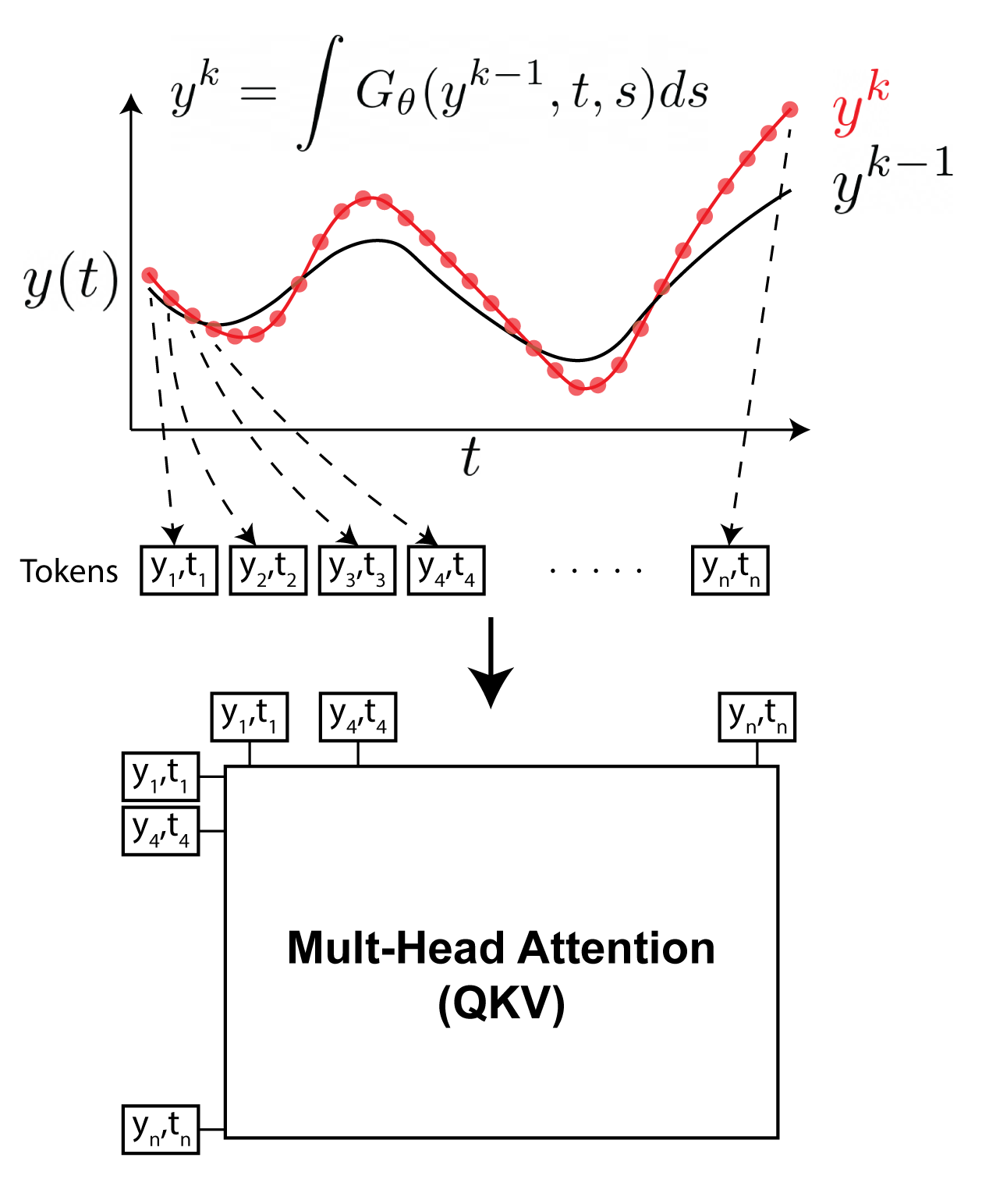
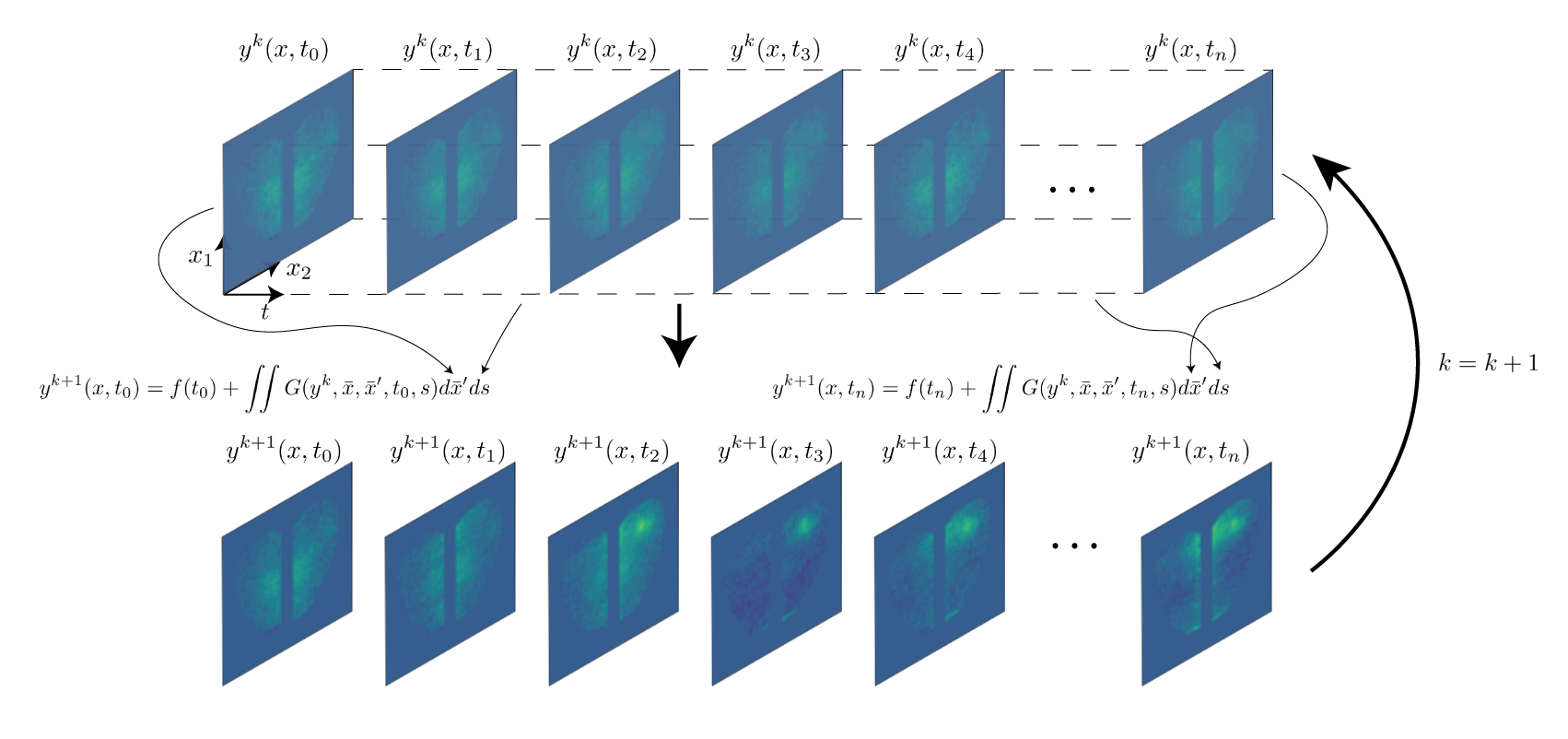
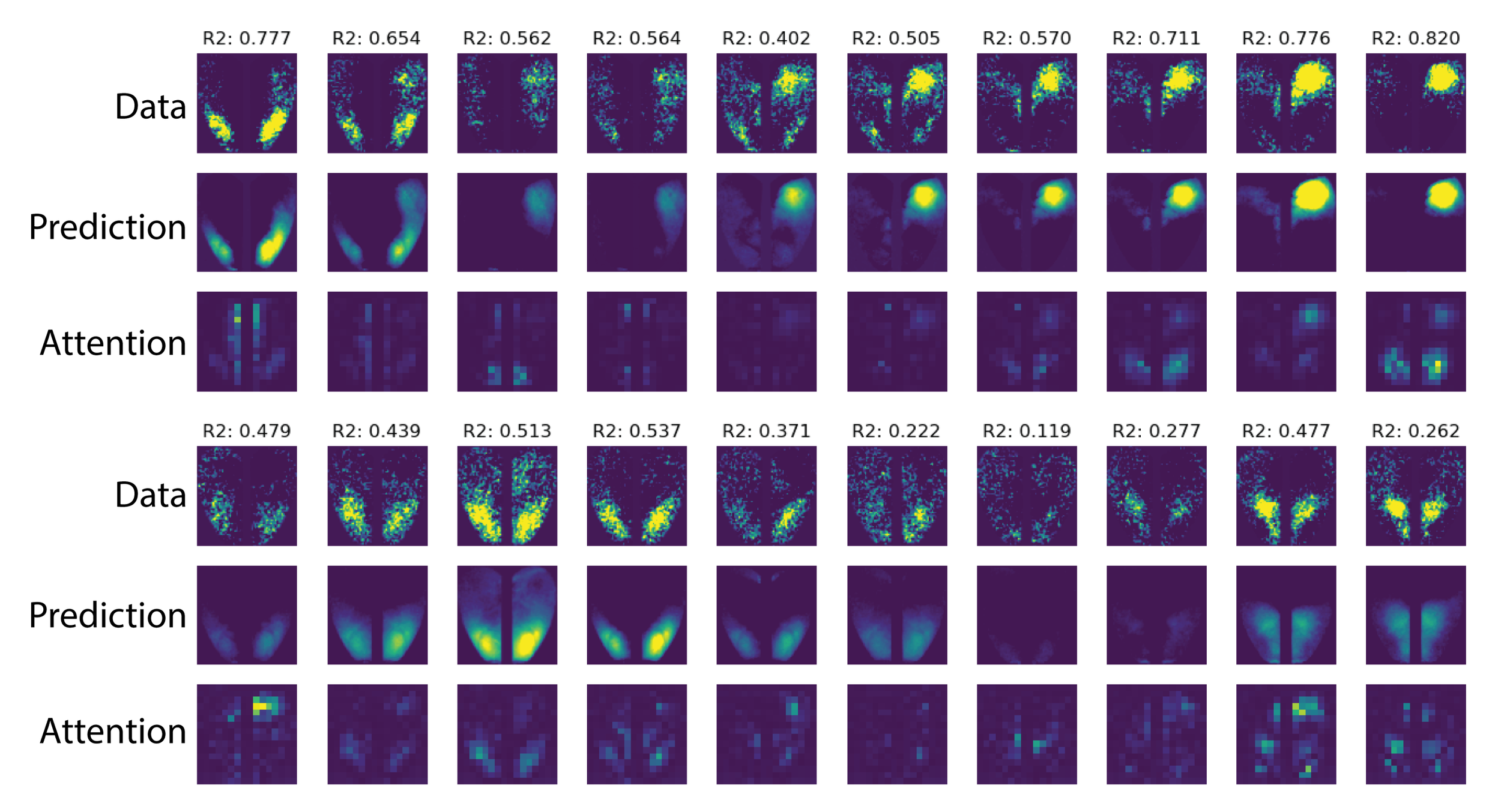
Neural Integral Equations
Neural Integral Equations (NIE) and Attentional Neural Integral Equations (ANIE) learn complex spatiotemporal dynamics via differentiable integral‑equation solvers, treating operator learning as an optimization problem. They outperform neural ODEs on ODE/PDE tasks and provide interpretable embeddings and attention maps for systems like brain‑activity recordings.
scGAT: Single-Cell Graph Attention Networks for Interpretable Identification of Cell State Determinants
scGAT applies graph attention to scRNA‑seq data to pinpoint transcripts driving distinct cell states, enabling biomarker discovery and drug‑repurposing hypotheses. It complements traditional differential‑expression pipelines with an interpretable, attention‑based framework.
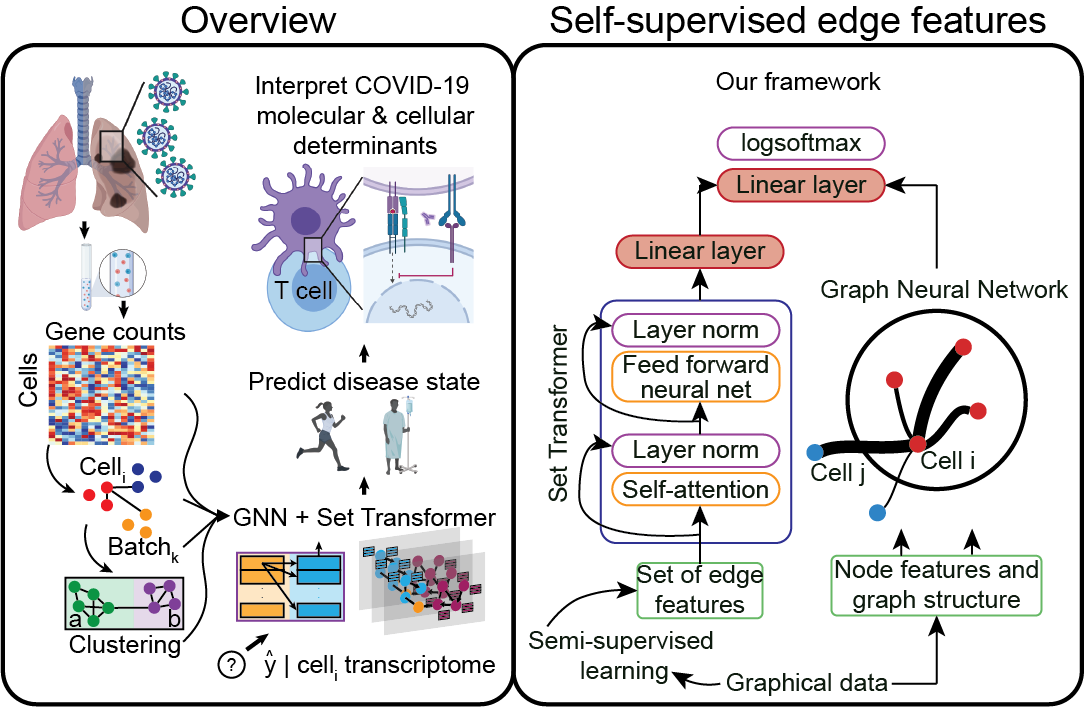
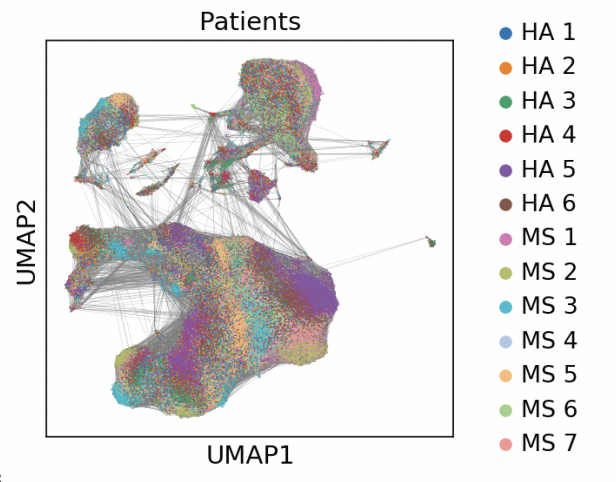
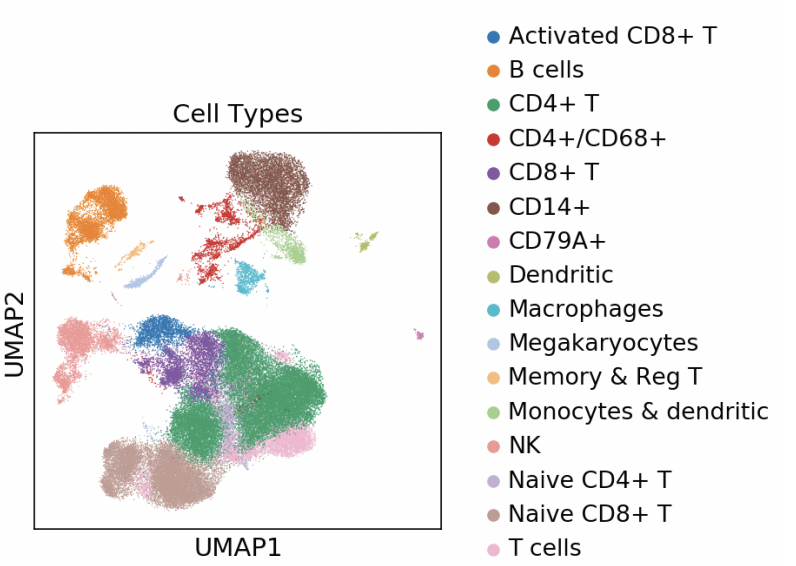
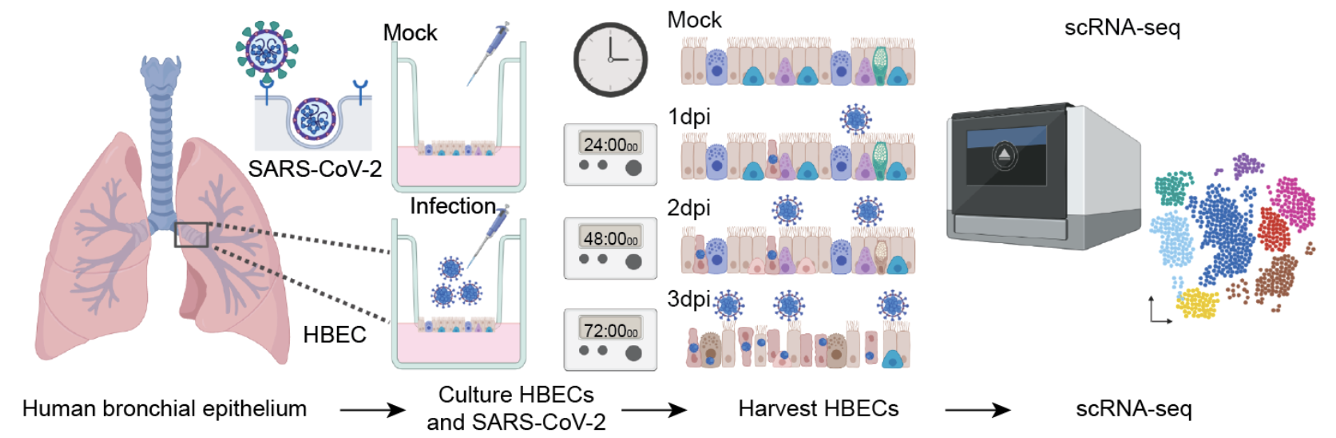
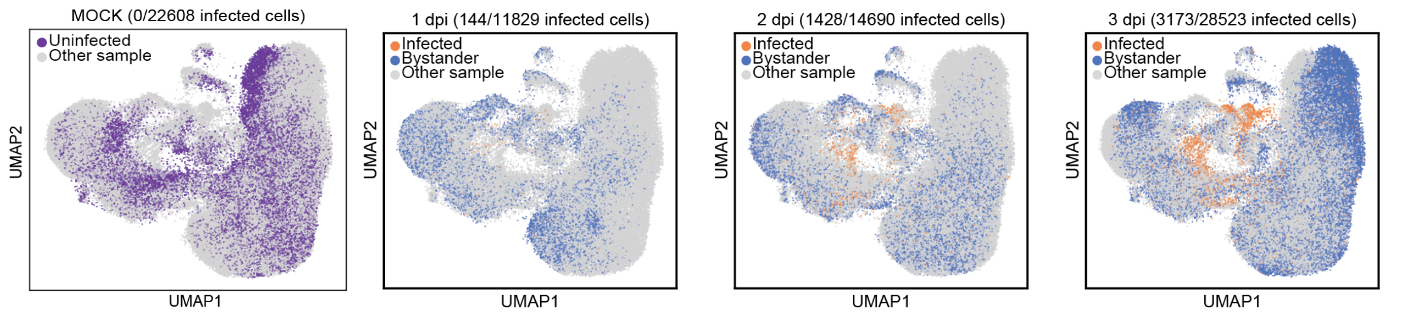
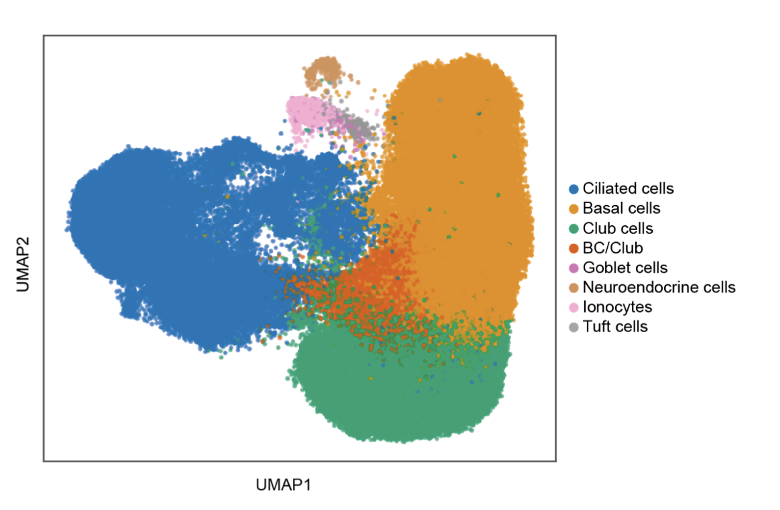
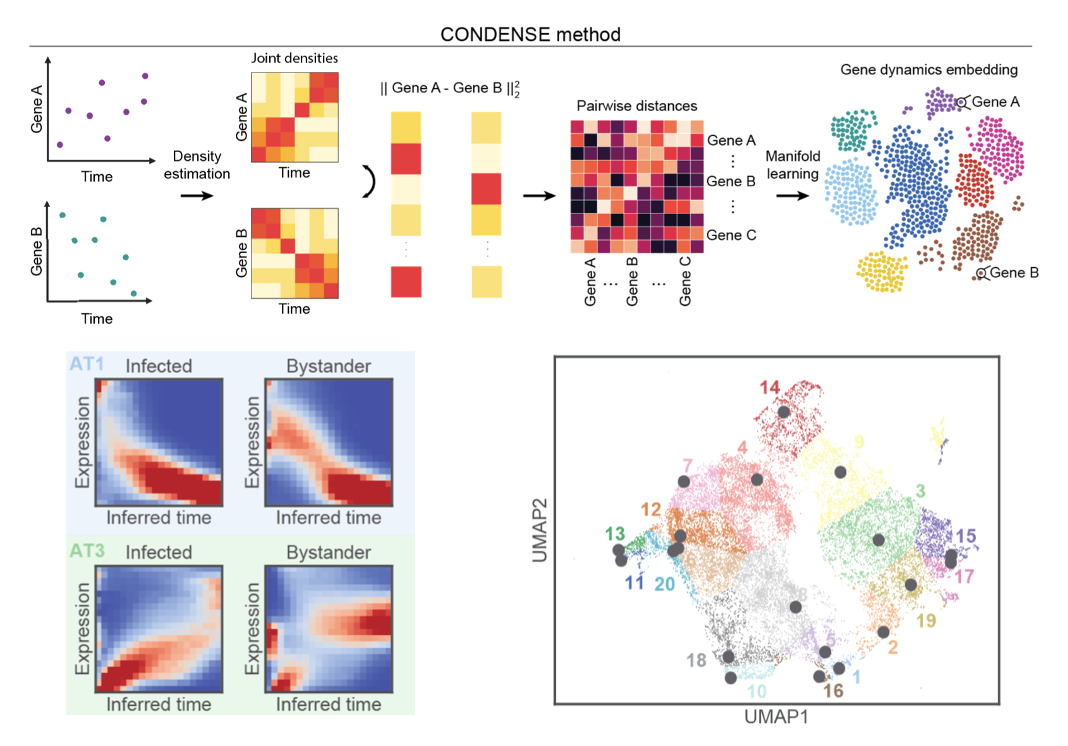
Single-cell Longitudinal Analysis of SARS-CoV-2 Infection in Airway Organoids
By profiling human bronchial epithelial cells (HBECs) over time in air-liquid interface cultures, this study uncovered novel polyadenylated viral transcripts, tracked expanding cell tropism of SARS-CoV-2, and mapped interferon‑stimulated responses in both infected and bystander cells. Electron and immunofluorescence microscopy validated ciliated cells as initial infection targets.
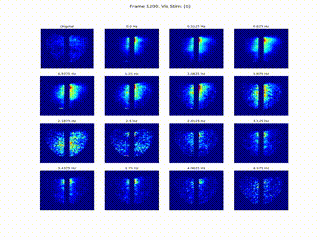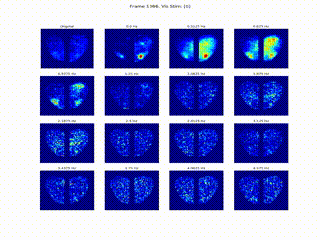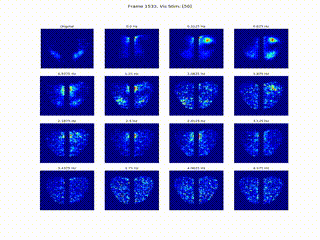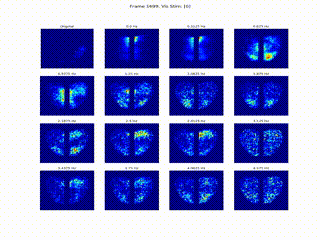
Inferring Neural Dynamics From Calcium Imaging Data Using Deep Learning
We develop continuous spatiotemporal ML frameworks—combining transformers and Fourier features—to model traveling waves in cortex calcium imaging data. This approach aims to reveal how neural waves are generated and propagate across brain networks.
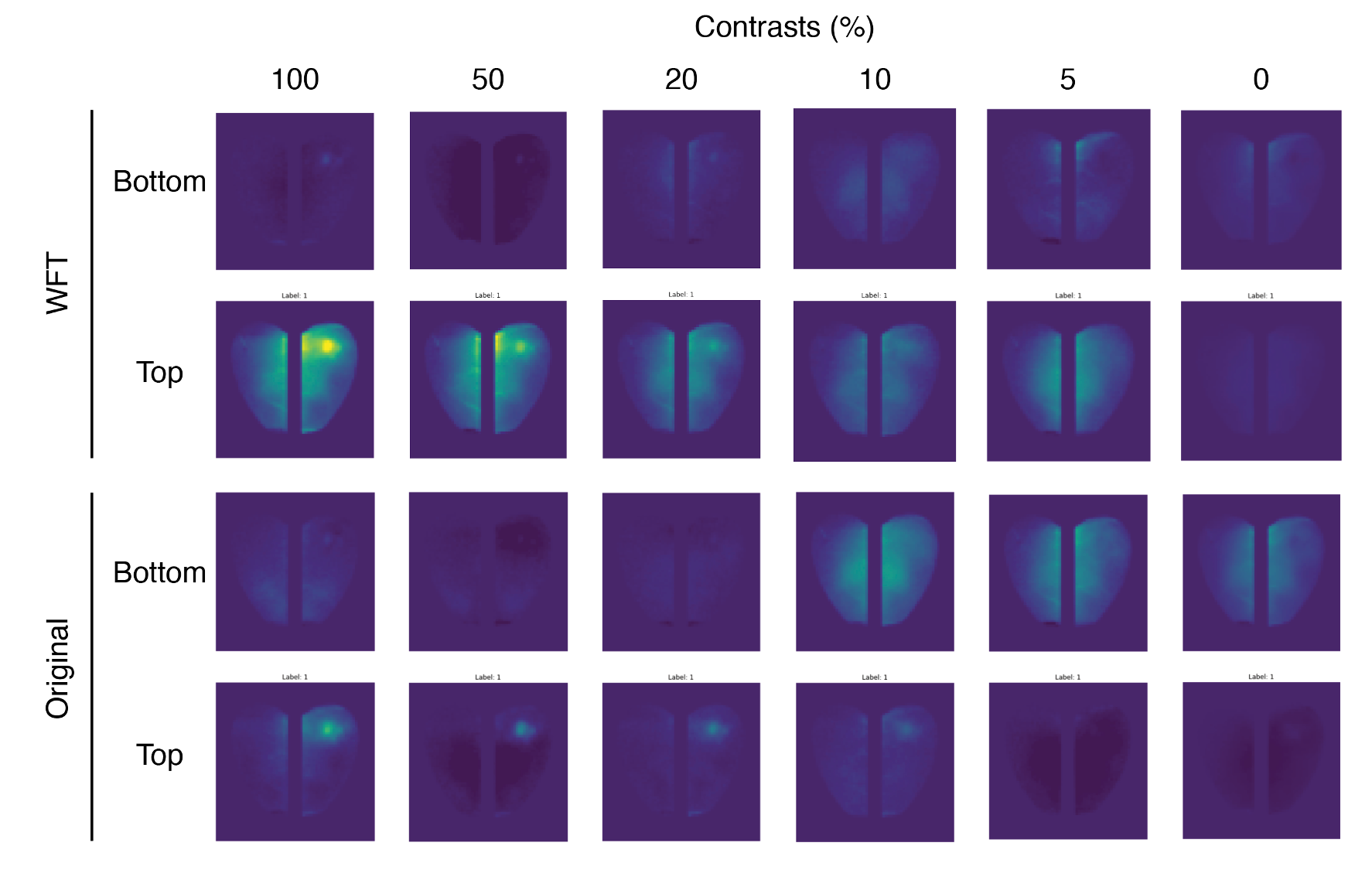
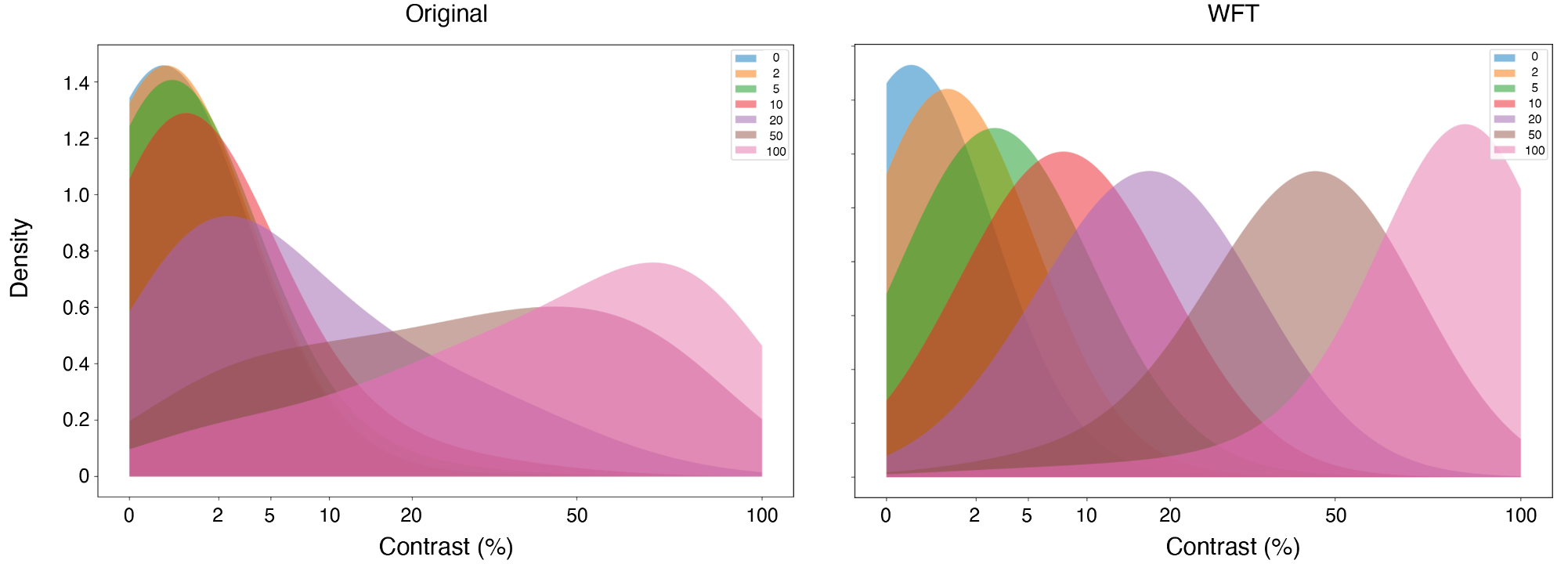
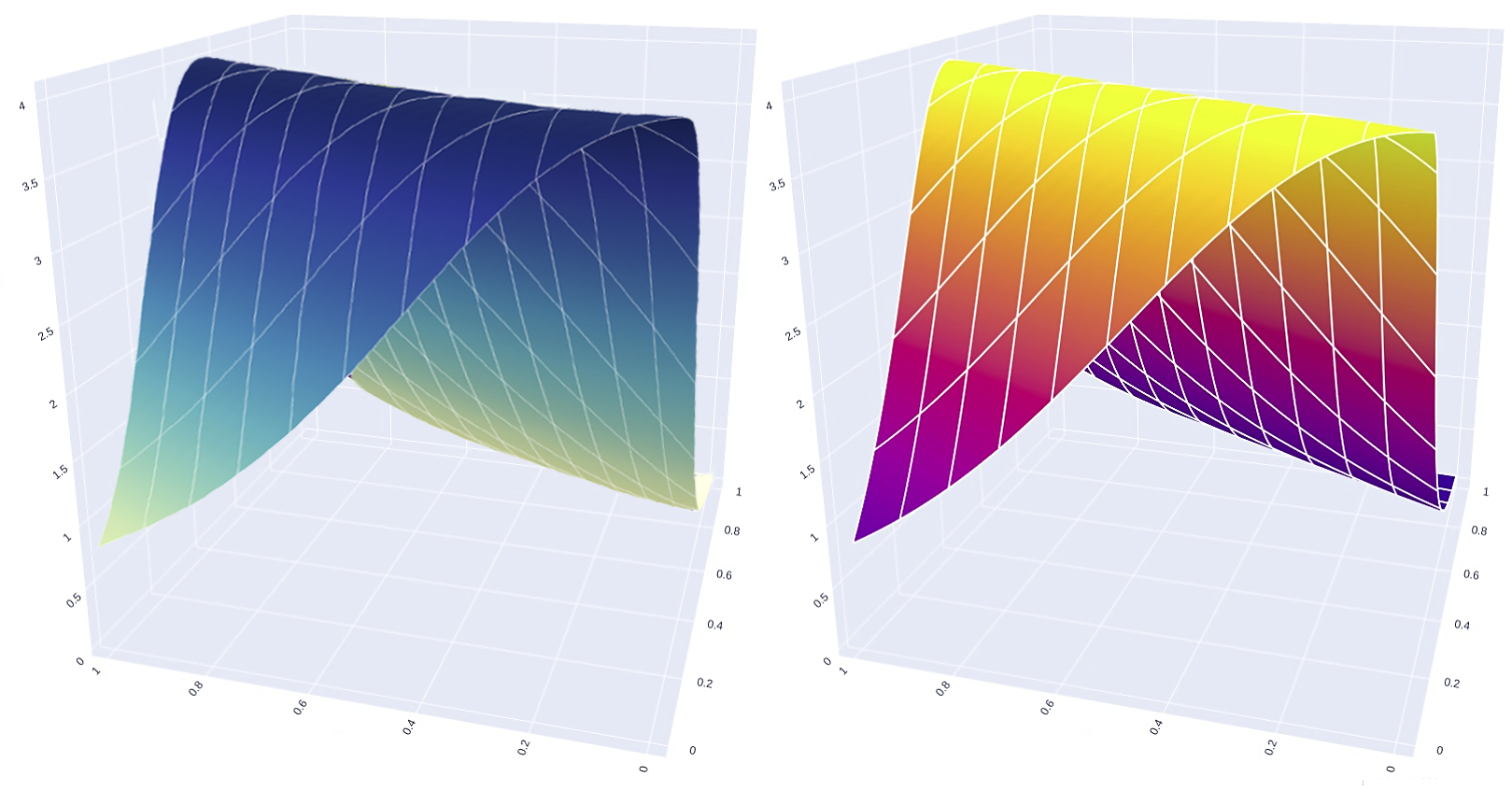
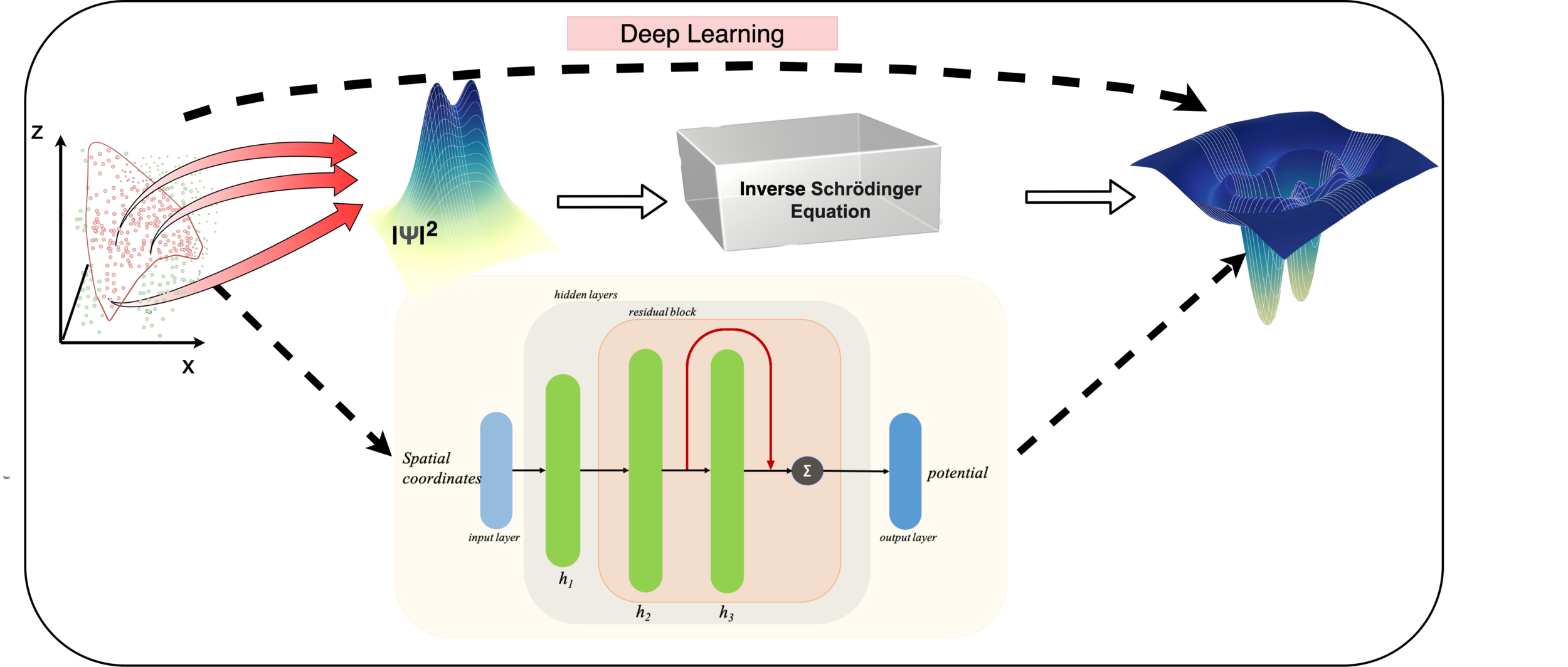
Learning Potentials of Quantum Systems using Deep Neural Networks.
Quantum Potential Neural Network (QPNN) approximates a system’s Hamiltonian by solving the inverse Schrödinger problem in an unsupervised manner, working across 3D and multi‑electron quantum systems.
Homology free functional annotation of enzymes
Leveraging graph neural networks, this project predicts enzyme function without sequence homology by learning features from small‑molecule and protein graph representations—unlocking insights into the “microbial dark matter” of unannotated genes.